Full HTML
Evaluation of multi-drug resistant isolates associated with clinical infections from two hospitals in the Dhaka region, Bangladesh
Mahmudul Hasan Masud1, Md. Sajedur Rahman1, Nusrat Jahan1, Atikur Rahman Titas1, Zakaria Ahmed Sany1, Fahim Faysal1, Umma Hany Sawon2, Md. Aoulad Hosen1,3
Author Affiliation
1Dept. of Microbiology, Gono Bishwabidyalay, Savar, Dhaka, Bangladesh
2Dept. of Pharmacy, Comilla University, Comilla, Bangladesh
3Dept. of Microbiology, Faculty of Veterinary and Animal Science, Hajee Mohammad Danesh Science & Technology University,
Dinajpur, Bangladesh
Abstract
Background: The emergence of bacterial antibiotic resistance represents a significant global health concern, particularly with the rise of multi-drug-resistant bacteria. Overuse of antibiotics has been identified as a primary driver of this phenomenon, as exposure to these drugs can lead to developing resistance in microorganisms. This study aims to investigate the incidence of MDR-resistant isolates in clinical samples (blood, urine and pus) collected from patients at Gonoshasthya Somaj vittik Medical College & Hospital and LABAID Hospital, Dhaka, Bangladesh.
Materials and Methods: The research was conducted for 12 months from July 2022 to June 2023.Cultural and biochemical tests were performed to isolate and identify the bacterial isolates from different clinical samples. Antibiograms were determined using the disc diffusion method with ten commercially available broad-spectrum antibiotics.
Results: A total of 146 isolates including Gram-positive, Gram-negative and fungal strains were isolated and identified through standard biochemical tests. We recorded 37.73% MDR E. coli and 55.55% Staphylococcus spp. In this research, where the highest number of MDR E. coli found in urine samples was 45% and MDR Staphylococcus spp. Found 66.67% in the pus sample of examined patients, respectively. The antibiotic susceptibility testing revealed significant resistance patterns among the isolates. E. coli showed high resistance rates in Azithromycin 100%, followed by Aztreonam 90%, and Amoxyclave 75%, whereas Staphylococcus spp. notably resistance in Penicillin-G and Cefixitin at 100% followed by Tetracycline at 86.67% and Erythromycin at 73.33% respectively.
Conclusion: These findings underscore the urgent need for effective management strategies due to high antibiotic resistance in both Gram-positive and Gram-negative bacteria. Understanding bacterial resistance patterns is essential for guiding treatment and infection control in clinical settings. Our research highlights the immediate need to discover new antibiotics for treating human multi-drug resistant (MDR) infections.
DOI: 10.18231/j.yjom.2024.007
Keywords: Multi-drug Resistant, Gram-Negative Bacteria, Gram-Positive Bacteria, Disc Diffusion method
Pages: 114-121
View: 4
Download: 10
DOI URL: http://doi.org/10.18231/j.yjom.2024.007
Publish Date: 11-09-2024
Full Text
Antibiotic resistance (ABR) represents a multifaceted global health crisis fueled by a complex interplay of factors, including overuse and misuse of antibiotics, inadequat healthcare infrastructure, and socio-economic disparities. While ABR poses a formidable challenge worldwide, its impact is particularly pronounced in low- and middleincome countries like Bangladesh. This introduction aims to provide a comprehensive overview of the ABR landscape in Bangladesh, drawing on existing literature to elucidate key drivers, challenges, and implications of antibiotic resistance in the country. Bangladesh, a densely populated country in South Asia, grapples with many healthcare challenges, including limited access to quality healthcare services, widespread poverty, and inadequate sanitation infrastructure. Against this backdrop, the misuse and overuse of antibiotics have emerged as major contributors to the escalating burden of ABR. In resource-constrained settings, antibiotics are often prescribed indiscriminately, without proper diagnostic confirmation or adherence to treatment guidelines. 1 Moreover, the availability of antibiotics without prescription, coupled with a lack of regulatory oversight, exacerbates the problem of antibiotic misuse among the general population. 2 Studies conducted across various healthcare settings in Bangladesh have documented alarmingly high rates of antibiotic resistance among clinically significant pathogens. For instance, a study by3 reported widespread resistance to commonly used antibiotics such as ampicillin, ciprofloxacin, and co-trimoxazole among clinical isolates of Escherichia coli and Klebsiella pneumonia. Similarly, research conducted by4 highlighted the emergence of multidrug-resistant strains of Acinetobacter baumannii in hospital settings, posing significant challenges for infection control and patient management. Despite the growing recognition of ABR as a public health threat, surveillance efforts in Bangladesh still need to be completed and adequate. As stated by Haque et al., 5 the absence of standardized surveillance protocols, limited laboratory capacity, and inadequate data-sharing mechanisms impede the timely detection and response to emerging resistance patterns. Furthermore, the need for coordination among stakeholders, including healthcare providers, policymakers, and researchers, hampers the development of comprehensive strategies to address ABR. 6 The repercussions of antibiotic resistance extend beyond individual patient outcomes, encompassing broader socioeconomic and public health implications. Treatment failures resulting from antibiotic resistance not only increase healthcare costs but also contribute to prolonged hospitalizations and higher mortality rates. 7 Moreover, the spread of resistant pathogens within healthcare facilities and communities amplifies the risk of outbreaks and undermines the effectiveness of infection control measures. 8 In conclusion, Bangladesh’s escalating threat of antibiotic resistance underscores the urgent need for concerted action to address this complex public health challenge. Efforts to combat ABR must encompass multifaceted interventions, including enhanced surveillance systems, rational antibiotic use policies, and investments in healthcare infrastructure. By adopting a comprehensive and collaborative approach, Bangladesh can mitigate the impact of antibiotic resistance and safeguard the health and well-being of its population. Therefore, this study aims to isolate and identify multi-drug resistant isolates from various clinical samples collected in different hospitals in Dhaka, Bangladesh and to assess the multi-drug resistance profiles of the isolated bacteria.
Study design and period
The design followed in this research is a descriptive crosssectional study. The study period of the research was from July 2022 to June 2023.
Sampling
A total of 100 human clinical samples were collected from Gonoshasthya Somajvittik Medical College Hospital (50) Savar and LABAID Hospital (50) Dhaka, Bangladesh. The clinical samples (blood, urine and pus) were collected from different age groups (11-74 years), including both male and female patients. Subsequently, all samples were properly labeled and transferred in an ice box to the laboratory of the Department of Microbiology, Gono Bishwabidyalay, for further processing and bacteriological analysis. The research commenced in December 2021 at the research laboratory of the Department of Microbiology, Gono Bishabidyalay, Dhaka, Bangladesh.
Isolation and identification of isolates
Samples were collected from clinically relevant areas, and data analysis was based on various parameters such as study period, location, clinical syndrome, age group, pathogen, susceptibility testing method, susceptibility testing standard, and source of infection. Bacterial isolation was performed using streaking principles, with subsequent identification based on colony morphology, Gram staining, and conventional biochemical tests. The collected samples were processed according to. 5 The specimens were cultured on Nutrient agar, Blood agar, MacConkey agar, EMB agar, SS agar, Cetrimide agar, Mannitol Salt Agar and Chromogenic agar, followed by overnight incubation at 37◦C in an incubator (BIOBASE, China).
Gram staining
Gram staining was employed to morphologically differentiate bacteria based on cell wall constituents as primary identification. Crystal violet, Gram’s iodine, ethanol or acetone, and safranin were utilized as stains. Gram-positive and Gram-negative bacteria were distinguished based on gram reactions.
Biochemical tests
Biochemical tests, including catalase, oxidase, coagulase, motility agar, urease, methyl red (MR), Voges-Proskauer (VP), and Triple Sugar Iron (TSI) tests were conducted for bacterial identification. Positive and negative reactions were interpreted based on specific colour changes or the presence of characteristic products. All media were purchased from HI Media Private Ltd., India. A loopful of each biochemically confirmed isolate was then inoculated into the nutrient broth containing 15% glycerol and frozen at -70°C for further use. All media were purchased from HI Media Private Ltd., India.
Antibiotic sensitivity test
All biochemically identified isolates (n=146) were tested for their susceptibility to different antibiotics. In vitro antibiotic susceptibility testing was performed using the standardized agar disk diffusion method. Muller Hinton agar was employed, and the potencies of ten antibiotics were tested against various bacterial isolates. The tested antimicrobials discs were Azithromycin (15µg), Aztreonam (30µg), Amoxyclave (20µg), Cefixime (5µg), Ceftazidime(30µg), Ampiciline (10µg), Cefuroxime (30µg), Ciprofloxacin (5µg), Cotrimoxazol (25µg), and Gentamycin (10µg). The antibiotic sensitivity test was determined according to Clinical and Laboratory Standard Institute (CLSI) guidelines for agar disk diffusion (CLSI, 2021). 9 A pure bacterial colony (0.1 ml) was spread on the MuellerHinton agar plate, and antibiotic discs were placed on the plates. Then, the plates were incubated at 37◦C for 16 to 18 hours in a standard incubator (BIOBASE, china). After overnight incubation, the inhibition zones were measured using a millimetre scale according to9 guidelines. All diLegends: LAH: LABAID Hospital, GSVMC: Gonoshasthya Somajvittik Medical College were purchased from HI Media Private Ltd., India. All tests were done three times for result accuracy.
Multidrug Resistance (MDR) Strains Identification
Multi-drug resistance bacterial isolates were determined by using the disc diffusion method, according to. 10 Multi-drug resistance (MDR) was taken as resistant to four or more antibiotics tested.
Multiple Antibiotic Resistance (MAR) index determination
According to, the Multiple Antibiotic Resistance (MAR) index for each strain was determined, which, MAR index= No. of resistance isolates / Total number of tested antibiotics MAR index= A/B Where,
No of resistance isolates
B=Total number of tested antibiotics;
Statistical analysis
Statistical analysis was done based on data variables of isolates. All data were inputted on an Excel sheet, and then Excel version 21(different formulation) was applied for data analysis.
Ethical Approval
The ethical approval of the current research was taken from the hospital administrative office.
Association between frequency of isolates and patients
A total of 146 bacterial isolates were isolated and identified out of 55 positive cases from different sources. In this research, based on gender, a higher frequency of isolates was observed in females 91 (62.33%) whereas in males 55 (37.67%) respectively (Table 1). Regarding the age of patients, the result revealed that the highest number of isolates found in the 31-60 years age group, with 54 (36.99%) and 40 (27.40%), were found in the age group 61-74 years, respectively. Concerning the type of clinical samples, a high frequency of isolates, 79 (54.11%) observed in urine compared to other samples (Table 1). According to the sampling area, 86 (56.16%) isolates were identified in Gonosastho Hospital compared to Lab Aid Hospital 64 (43.84%) which was presented in (Table 1).
Frequency of isolates with blood, urine, and pus sample
In this study, out of 53 E. coli isolates, the highest number of isolates were recovered from urine sample 28 (52.83%), followed by pus sample 13 (24.52%) and blood sample 12 (22.64%) respectively, whereas Klebsiella spp. was measured 24 (66.67%) in urine and the least number found in blood and pus 6 (16.67%) (Table 2). Additionally, we recovered 27 (100%) Candida spp. In urine samples from female patients (Table 2). According to our study, A total of 146 isolates were isolated and identified by different cultural and biochemical tests, including the highest prevalence of Gram-negative bacteria, 92 (64%), whereas Gram-positive bacteria, 27 (18%) and fungal strain 27(18%), respectively (Figure 1). In a blood sample, we recovered 32 isolates with the highest number of E. coli obtained 7/12 (58.33%) in the age group 31-60 years, Staphylococcus spp. 6/11 (54.54%) in the age group 31-60 years, Klebsiella spp. found 3/6 (50%) in the age group 11-30 years, whereas no Candida spp. was found (Figure 2). Consequently, in urine samples, 79 bacterial strains were recovered where 10/28 (35.71%) E. coli were found in age group 31-60 years and 61- 74 years, 9/24 (37.50%) Klebsiella spp. in 61-74 years, 10/27 (37.03%) Candida spp. found in the age group 11- 30 years and 9/27 (33.33%) in age 61-74 years respectively (Figure 3). In this research, a total of 35 isolates were obtained from pus samples, including the highest 7/16 (43.75%) Staphylococcus spp. in the age group 11-30 years, 5/13 (38.46%) E. coli in the age group 11-30 years and 31- 60 years respectively (Figure 4).
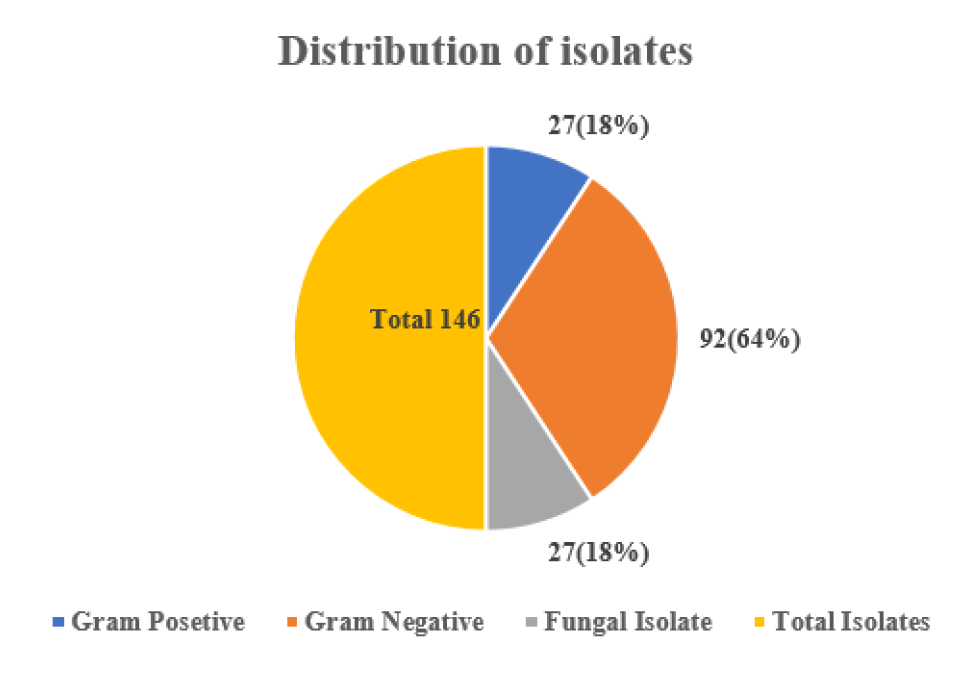
Figure 1: Frequency distribution of isolates in human clinical samples
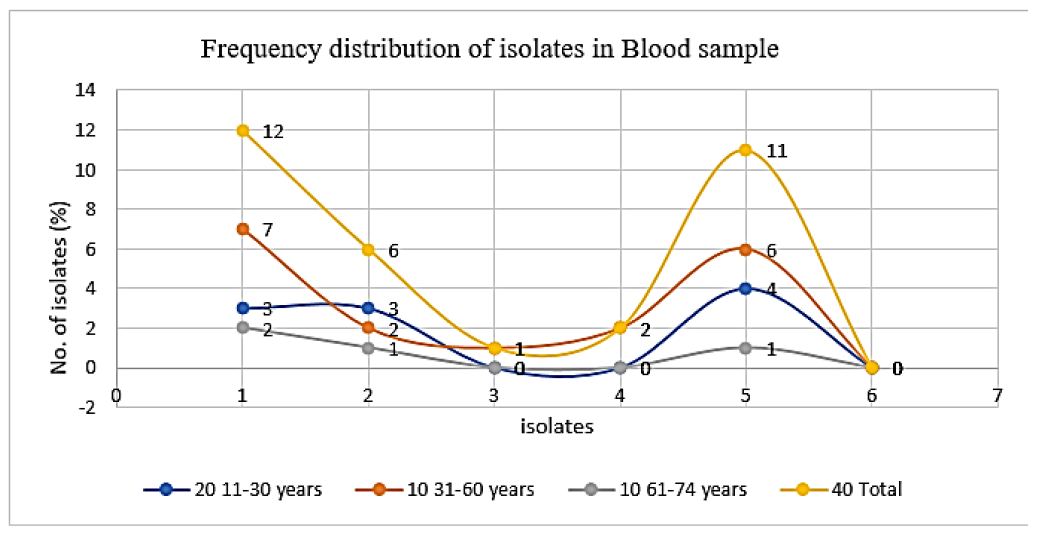
Figure 2: Frequency distribution of isolates in blood samplesbased on age group 1: E.coli, 2: Klebsiella spp., 3: Salmonella spp., 4: Pseudomonasspp., 5: Staphylococcus spp., 6: Candidaspp.

Figure 3: Frequency distribution of isolates in urine samplesbased on age group 1: E.coli, 2: Klebsiella spp., 3: Salmonella spp., 4: Pseudomonasspp., 5: Staphylococcus spp., 6: Candidaspp.

Figure 4: Frequency distribution of isolates in pus sample based on age group 1: E.coli, 2: Klebsiella spp., 3: Salmonella spp., 4: Pseudomonasspp., 5: Staphylococcus spp., 6: Candidaspp.
Antibiotic resistance pattern and MAR index calculation
In the present study, out of 146 isolates, 35 (24%) multidrug resistant bacteria were found. According to the results, 20/146 (14%) multi-drug resistances E. coli were identified, and resistance profiles were shown on different commercial antibiotics. E. coli demonstrated high resistance toward azithromycin 20 (100%), followed by aztreonam 18 (90%), amoxiclav 15 (75%), and ciprofloxacin 11 (55%) compared to other commercial antibiotics (Table 3). Notably, this research measured the least resistance profile of ceftazidime 2 (10%) in blood and pus samples from Lab Aid Hospital. Based on CLSI interpretation criteria, 37.73% (20/53) of multidrug-resistant E. coli were mentioned in this study. Consequently, 45% (9/20) of multidrug-resistant E. coli were found in the urine sample, followed by 35% (7/20) in the pus sample and 20% (4/20) in the blood sample, respectively. Table 3 represents the MAR index calculation of isolated multi-drug resistance E. coli from different sources. In tabLegends: LAH: LABAID Hospital, GSVMC: Gonoshasthya Somajvittik Medical College 3, the MAR index ranged from 0.3 to 0.8, and the average MAR index was 0.43. Additionally, the highest number of antibiotics showed resistance (MAR index 0.8) in blood samples from Lab Aid Hospital, Dhaka, compared to other samples as well as another hospital. According to the obtained results, out of 27 Staphylococcus spp. isolates, 15 multi-drug rLegends:
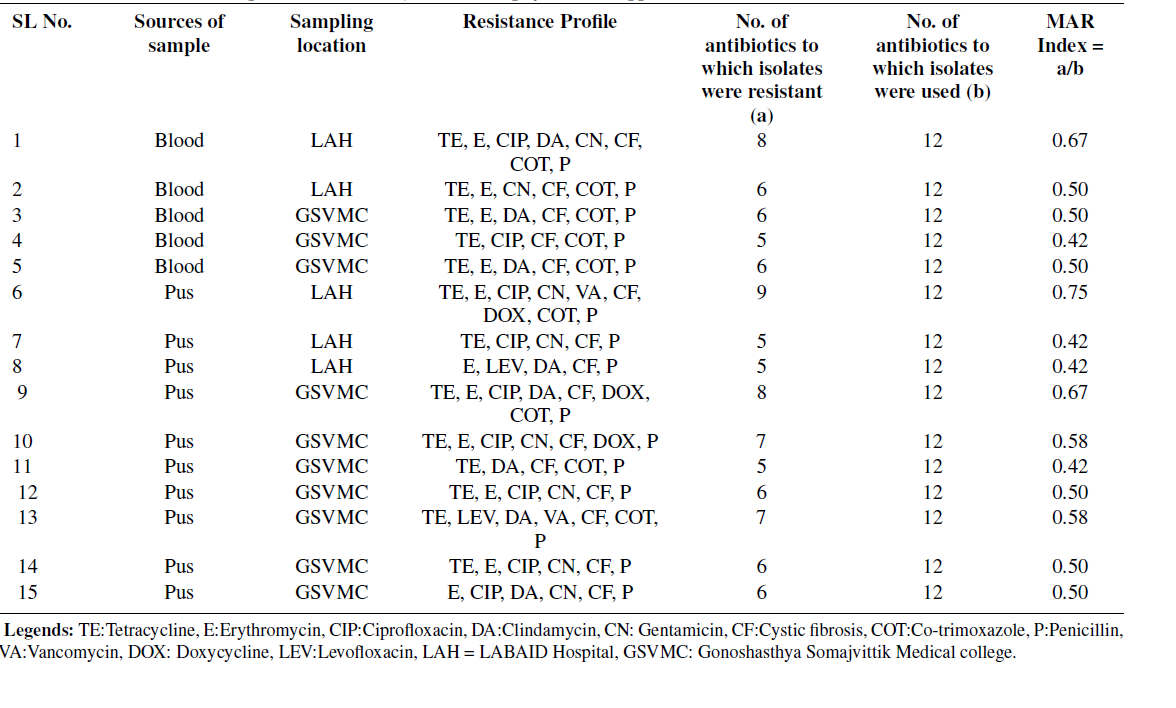
LAH: LABAID Hospital, GSVMC: Gonoshasthya Somajvittik Medical college resistance Staphylococcus spp. were identified and showed resistance profile on different commercial antibiotics. Staphylococcus spp. showed high resistance to penicillin-G and cefixiLegends: LAH: LABAID Hospital, GSVMC: Gonoshasthya Somajvittik Medical College 15 (100%), followed by tetracycline 13 (86.67%), erythromycin 11 (73.33%), ciprofloxacin and cotrimoxazole 9 (60%) compared to other commercial antibiotics (Table 4). Notably, in this research, the least resistance profile of vancomycin 1 (6.67%) was measured in pLegends: LAH: LABAID Hospital, GSVMC: Gonoshasthya Somajvittik Medical collegeus sample from Gonosastho Hospital. Based on CLSI interpretation criteria, 55.55% (15/27) multi-drug resistance Staphylococcus spp. were mentioned in this study. Consequently, 66.67% (10/15) of multidrug-resistant Staphylococcus spp. were found in pus samples, followed by 33.33% (5/15) in blood samples, respectively. Table 4 represents the MAR index calculation of isolated multi-drug resistance Staphylococcus spp. from different sources, whereas the MAR index ranged from 0.42 to 0.75, and the average MAR index was 0.52. Additionally, the highest number of antibiotics showed resistance (MAR index 0.75) in the pus sample from Lab Aid Hospital, Dhaka, compared to other samples as well as another hospital.
Table 1: Association of isolates with patient’s demographic characteristic (Gender, Age Group, Sampling type and area)
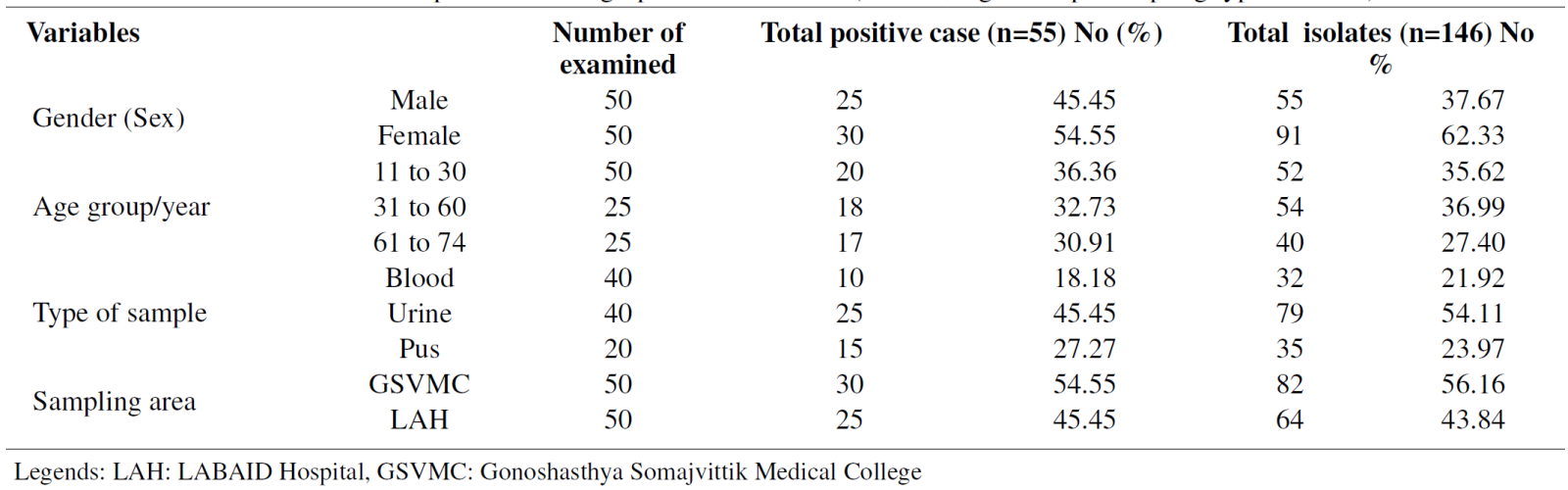
Table 2. Frequency distribution of isolates with patient samples (Blood, Urine and Pus)
Table 3. Antibiotic Resistance profile of Multidrug-resistantE. coli from different sources
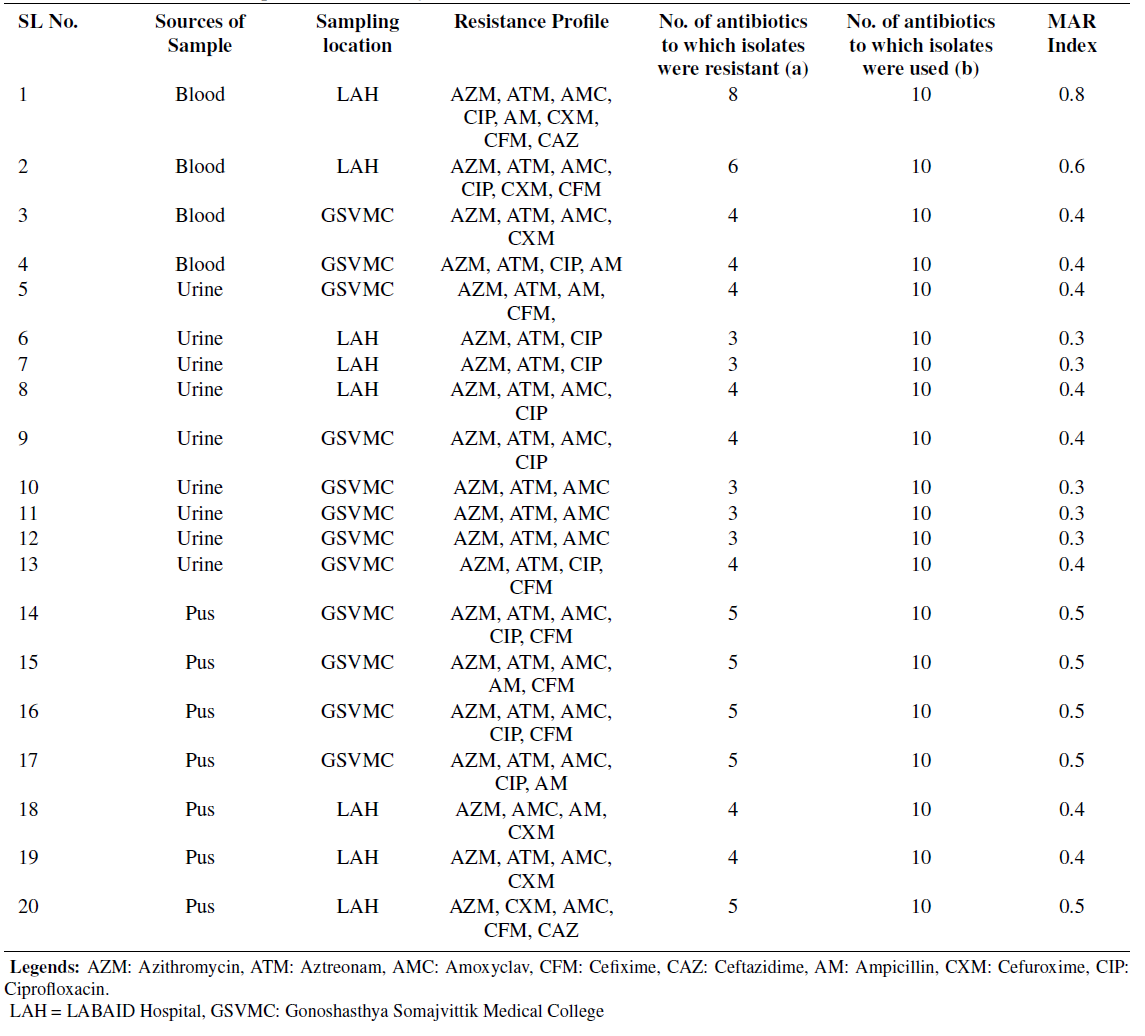
Table 4: Antibiotic resistance profile of multidrug-resistantStaphylococcus spp. from different sources
The escalating issue of multi-drug resistance (MDR) in bacteria poses a significant challenge for clinicians, given the ability of these microorganisms to develop resistance to multiple antibiotics. This resistance not only results in a financial burden and increased treatment failures but also facilitates the rapid spread of infections from person to person. Over the past fifty years, the pervasive use of antibiotics has contributed to the emergence of non-fermentative Gram-negative bacteria as critical healthcare-associated pathogens. Consequently, recent studies have concentrated on identifying these bacteria and monitoring their susceptibility patterns to improve infection management strategies. Establishing the clinical relevance of isolated non-fermentative Gramnegative bacilli is crucial before classifying them as pathogens, which can prevent unnecessary antibiotic usage and the rise of drug-resistant strains. Our study addresses the limited data on the prevalence of MDR E. coli and Staphylococcus spp., highlighting their isolation from diverse clinical samples (blood, urine, and pus) in suspected patients aged 11-74 years. The highest number of MDR E. coli isolates was observed in the 31-60 age group, followed by the 11-60 and 61- 74 years groups.
Similarly, MDR Staphylococcus spp. Showed the highest prevalence in the 31-60 years age group. Gender-wise distribution revealed a higher incidence of MDR isolates in females (62.33%); these findings align with previous studies by Muhammad Zahid11, where the occurrence was lower in males (47%, n= 324) as compared to females (53%, n = 324). This trend may be attributed to social and cultural factors in developing countries like Bangladesh, where females often face neglect, leading to poorer food hygiene and weaker immune systems. Additionally, frequent hospital admissions due to childbirth increase their exposure to infections. In our study, we identified 79 bacterial strains, with notable findings such as 35.71% of E. coli (10/28) occurring predominantly in the age groups of 31-60 years and 61- 74 years, 37.50% of Klebsiella spp. (9/24) in the 61-74 age group, and 37.03% of Candida spp. (10/27).
These results closely align with those reported by Mohammad Aminul Islam12, who observed E. coli (51.6%) as the primary pathogen, followed by Streptococcus spp. (15.7%) and Klebsiella spp. (12.1%) in individuals aged ≥18 years. In the present study, out of 146 isolates, 35 (24%) were identified as multidrug-resistant bacteria. This percentage is considerably lower than the findings of Meng Wang13 , who reported that among 7579 bacterial isolates from patients with healthcare-associated infections (HAI), 3223 (42.5%) were multidrug-resistant. According to this study, 20 out of 146 (14%) isolates were identified as multidrugresistant E. coli, exhibiting resistance to various commercial antibiotics. In contrast, S. Shakya 14 found a significantly higher prevalence of multidrug-resistant E. coli in their study, with 1000 out of 1865 (54%) isolates showing resistance to commonly used antibiotics for urinary tract infections (UTIs). Based on CLSI interpretation criteria, 55.55% (15/27) of Staphylococcus spp. isolates in this study were identified as multidrug-resistant. Specifically, 66.67% (10/15) of these multidrug-resistant Staphylococcus spp. were found in pus samples, and 33.33% (5/15) were in blood samples. All Staphylococcus spp. isolates showed high resistance to penicillin-G and cefixitin (100%). This study can be compared to the findings of Prashant Adhikari 15, where among 7433 clinical samples analyzed, Staphylococcus aureus was recovered from 499 (6.71%), with a prevalence of MRSA at 26.4% (95% CI 21.6% to 30.4%). The major source of MRSA was pus, accounting for 71 (18.5%) of the cases. 5.1.
Limitations and future researh Several limitations warrant consideration, including the study’s retrospective design and potential biases in patient selection and sample collection. Future research should focus on prospective studies with larger sample sizes to validate these findings across different healthcare settings. Additionally, molecular techniques could provide insights into genetic determinants of resistance, aiding in targeted therapeutic approaches.
The presence of MDR E. coli and Staphylococcus spp. in clinical samples, including blood, urine, and pus, highlights a severe public health concern in Bangladesh, particularly due to the potential transmission of these pathogens through food and water. The study’s findings underscore that MDR isolates, especially prevalent in the 31-60-year-old age group, pose significant challenges to infection prevention, given the possible impact on immunity in this demographic. The rising threat of MDR bacteria necessitates the urgent discovery of new effective antibiotics to safeguard human health and the environment. Bangladesh’s healthcare system and global health settings must prioritize surveillance of drug resistance in microorganisms, particularly in clinical sectors. Public health awareness campaigns should focus on the prudent use of antibiotics and emphasize preventive measures prescribed by health professionals. By fostering a comprehensive understanding of antibiotic resistance and promoting responsible antibiotic usage, we can mitigate the spread of MDR bacteria and protect public health.
Source of Funding
None.
Conflict of Interest
None.
Acknowledgments
The authors want to special thanks to Gonosastho Samajvittik Medical College Hospital and LABAID Hospital for their valuable support during the collection of samples. The authors also want to give special thanks to the Department of Microbiology, Gono Bishwabidyalay, for providing their Laboratory facilities.
References
1. Sarker S, Nahar A. Antimicrobial susceptibility pattern of pathogenic bacteria in a tertiary care hospital in Dhaka, Bangladesh. Bangladesh J Infect Dis. 2018;5(1):6–10.
2. Sinha AK, Banik A. Antibiotic resistance pattern of clinical isolates of Escherichia coli in a tertiary care hospital in Dhaka City. J Inf P Health. 2018;11(2):241–2.
3. Hasan MR, Rahman M, Hasan MM. Antibiotic resistance patterns of Escherichia coli and Klebsiella pneumoniae isolated from urinary tract infections in a tertiary care hospital of Bangladesh. Int J Inf Dis. 2019;11(156):1–8.
4. Hossain A, Das RR, Islam MS. Antimicrobial resistance patterns of Acinetobacter baumannii in a tertiary care hospital in Dhaka. J Inf in Dev Count. 2020;14(4):400–4.
5. Haque A, Rahman MA, Molla MM. Antibiotic resistance of Escherichia coli isolated from food and clinical environment in China from 2001 to 2020. Sci Total Environ. 2018;10:173498.
6. Islam S, Hossain B, Rahman M. Antibiotic resistance pattern of bacterial isolates in intensive care unit of tertiary care hospital in Bangladesh. J Inf Pub Health. 2019;12(1):2–3.
7. Rahman M, Haque M. Prevalence of multidrug-resistant bacterial pathogens in a tertiary care hospital in Dhaka. J Inf Dev Count. 2017;36:491–6.
8. Rahman M, Haque M, Islam S. Bacterial profile and antimicrobial susceptibility pattern of blood culture isolates among septicemia patients in Dhaka. J Inf and Pub Health. 2019;17(1):1–15.
9. CLSI supplement M100). Clinical and Laboratory Standards Institute; 2021. Available from: https://clsi.org/media/1469/m100s27_sample. pdf.
10. Ezekiel CN, Olarinmoye AO, Oyinloye J. Distribution, antibiogram and multidrug resistance in Enterobacteriaceae from commercial poultry feeds in Nigeria. Afr J Microb Res. 2011;5(3):294–301.
11. Zahid M, Akbar M, Sthanadar AA. Isolation and identification of multi-drug resistant strains of non-lactose fermenting bacteria from clinical refuse in major hospitals of Khyber Pakhtunkhwa, Pakistan. J Med Microb. 2014;4(2):766–891.
12. Islam MA, Islam MR, Khan R. Prevalence, etiology and antibiotic resistance patterns of community-acquired urinary tract infections in. PLoS One. 2022;17(9):e0274423.
13. Wang M, Wei H, Zhao Y. Analysis of multidrug-resistant bacteria in 3223 patients with hospital-acquired infections (HAI) from a tertiary general hospital in China. Bosnian J B Med Sci. 2019;19(1):86–93.
14. Shakya S, Edwards J, Gupte HA. High multidrug resistance in urinary tract infections in a tertiary hospital. Pub Health Action. 2021;11(1):24–31.
15. Adhikari P, Basyal D, Rai JR. Prevalence, antimicrobial susceptibility pattern and multidrug resistance of methicillin-resistant Staphylococcus aureus isolated from clinical samples at a tertiary care teaching hospital: an observational, cross-sectional study from the Himalayan country. BMJ Open. 2023;13(5):e067384.
